Monarch® HMW DNA Extraction Kit for Tissue
Product information| Code | Name | Size | Quantity | Price | |
|---|---|---|---|---|---|
T3060S |
Monarch HMW DNA Extraction Kit for Tissue |
5 preps | - | Unavailable in your region | |
T3060L |
Monarch HMW DNA Extraction Kit for Tissue |
50 preps | - | Unavailable in your region |
Monarch® HMW DNA Extraction Kit for Tissue
Catalog #T3060
Product Introduction
- Designed for extraction for long-read sequencing applications from a variety of tissues, bacteria, yeast, insect, amphibian, and more. See related Monarch Genomic DNA Purification Kit (NEB #T3010) for gDNA extraction for general NGS and sequencing (Illumina® and other).
- Isolate high molecular weight DNA (100 kb to megabase range) from soft organ tissues and bacteria and more for Oxford Nanopore®, PacBio® sequencing
- Uses unique, innovative technology for fast, user-friendly workflows, only 90 minutes for extraction compared to hours or days.
- Includes RNase A, Proteinase K, and microtube pestles, for effective RNA removal, digestion of cellular proteins, and sample homogenization.
- Obtain best-in-class yields of highly-pure and intact DNA
- For HMW gDNA extraction from cells and blood for long-read sequencing, see related Monarch HMW DNA Extraction Kit for Cells & Blood (NEB #T3050)
Featured Resources:
- See how the Monarch HMW DNA workflow significantly reduced workflow time in the Cas9 targeted sequencing approach in our joint Technical Note with The Jackson Laboratory.
- Check out our publication in Biotechniques outlining a simple approach to accurate quantitation of ultra high molecular weight (UHMW) DNA.
- Review our technical note examining the use and comparison of a novel method to isolate genomic DNA from bacteriophages.
- Product Information
- Protocols, Manuals & Usage
- FAQs & Troubleshooting
- Citations & Technical Literature
- Quality, Safety & Legal
- Other Products You May Be Interested In
Product Information
Description
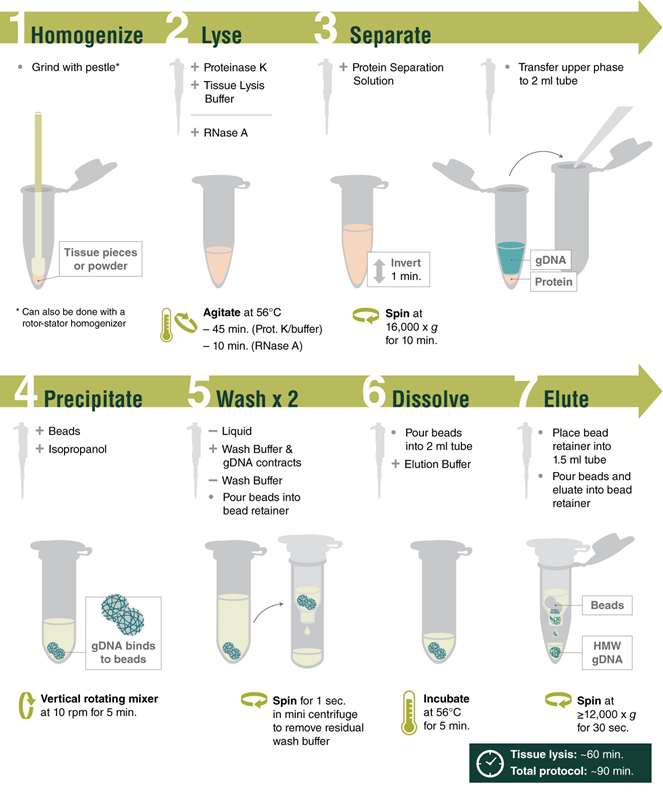
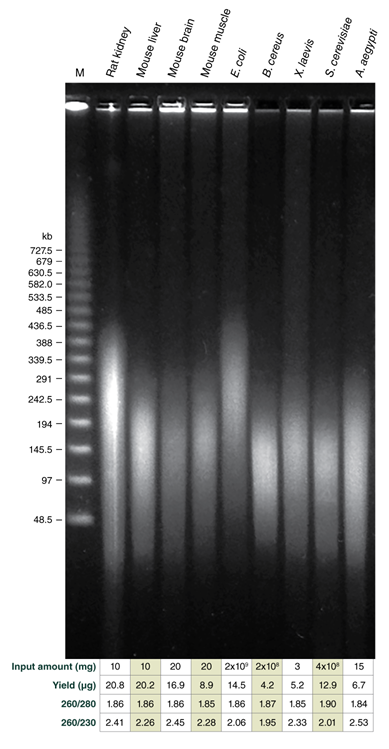
The Monarch HMW DNA Extraction Kit for Tissue provides a rapid and reliable process for extracting high molecular weight (HMW), intact genomic DNA from various tissues and bacteria, as well as other sample types including yeast, insect, and amphibian. The optimized extraction protocol for tissue utilizes pestle homogenization and proteinase K digestion with agitation for sample lysis, followed by a protein removal step and precipitation of the extracted DNA onto the surface of large glass beads. A slightly modified extraction protocol for bacteria utilizes lysozyme for the efficient lysis of the bacterial cell wall, prior to proteinase K digestion. DNA size ranges from 50- ≥500 kb for the standard protocols and into the Mb range when the lowest agitation speeds are used for processing soft organ tissues and bacteria. Purified DNA is recovered in high yield with excellent purity, including nearly complete removal of RNA. For tissue and bacteria, the processing time is 90 min. Purity ratios are typically 1.8-1.9 (260/280) for tissue and bacteria, 2.1-2.5 (260/230) for tissue, and 2.1-2.2 (260/230) for bacteria. Purified HMW DNA is suitable for a variety of downstream applications including long-read sequencing (Oxford Nanopore Technologies® and Pacific Biosciences®), optical mapping (Bionano Genomics®), and linked-read genome assembly.
Validated Sample Types
|
|
Visit our input guidelines for more information |
- This product is related to the following categories:
- Genomic DNA Extraction & Purification Products,
- Nucleic Acid Purification Products,
Properties & Usage
Related Products
Companion Products
Protocols, Manuals & Usage
Protocols
- Download Quick Protocol Card for Tissue
- Protocol for High Molecular Weight DNA (HMW DNA) Extraction from Tissue (NEB #T3060)
- Protocol for High Molecular Weight DNA (HMW DNA) Extraction from Bacteria (NEB #T3060)
- Protocol for High Molecular Weight DNA (HMW DNA) Extraction from Yeast (NEB #T3060)
- Protocol for High Molecular Weight DNA (HMW DNA) Extraction from Phage
- Protocol for UHMW DNA Cleanup in the Oxford Nanopore Technologies® UL Library Prep Workflow
- Protocol Guidance for Extraction of Ultra-High Molecular Weight (UHMW) Genomic DNA for Ultra-Long (UL) Read NGS Sequencing applications in Oxford Nanopore Technologies® workflows
Manuals
Usage & Guidelines
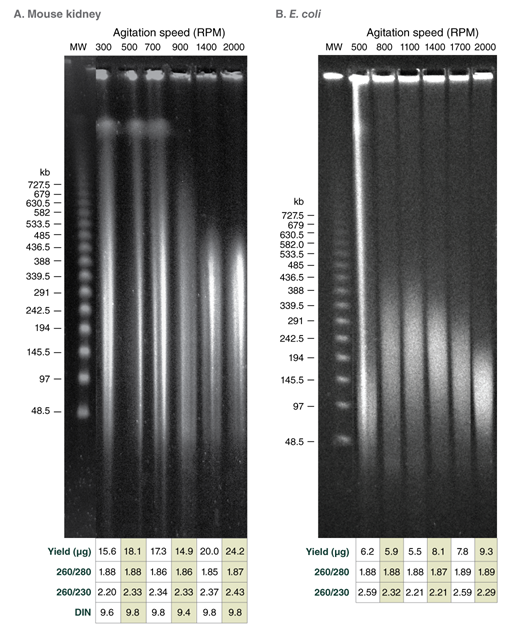
- Choosing an Agitation Speed During Lysis with the Monarch HMW DNA Extraction Kits
- Choosing Input Amounts for the Monarch HMW DNA Extraction Kits
- Considerations and Performance Data for Nanopore Sequencing of High Molecular Weight DNA (HMW DNA)
- Considerations for Loading Pulsed-Field Gels (PFG) with High Molecular Weight DNA
- Homogenization of High Molecular Weight DNA (HMW DNA) Samples After Elution
- Important Considerations for Starting Materials When Using the Monarch HMW DNA Extraction Kit for Tissues (NEB #T3060)
- Measuring, Analyzing & Storing High Molecular Weight DNA (HMW DNA) Samples
Application Notes
FAQs & Troubleshooting
FAQs
- How does excess residual protein in blood and tissue preps affect the purification process?
- Which measurement system is best used for assessing the concentration of HMW DNA?
- I dropped a few of my glass beads. Can I clean them, and do you supply extra?
- Is there anything special about the Monarch DNA Capture Beads, or can I use any glass beads?
- Are the Monarch HMW DNA Extraction kits compatible with samples stored in Monarch DNA/RNA Protection Reagent, RNAlater, DNA/RNA Shield etc.?
- How do I know that my HMW DNA has attached to glass beads in the Monarch HMW DNA Extraction Kits?
- Is there any benefit to adding in a size selection (e.g., Circulomics® Short Read Eliminator) following extraction?
- Can I still use the Monarch HMW DNA Extraction Kits if I do not have a thermal mixer?
- Can I use the Monarch HMW DNA Extraction Kits for processing plant samples?
- Can I use the Monarch HMW DNA Extraction Kits for processing insect samples?
- Can the Monarch HMW DNA Extraction Kit be used for processing yeast and/or fungal samples?
- Can the Monarch HMW DNA Extraction Kits be used to process marine samples, including marine invertebrates?
- Can the Monarch HMW DNA Extraction Kits be used to process human Buffy coat?
- Can the Monarch HMW DNA Extraction Kits be used to process mouthwash samples?
- Can the Monarch HMW DNA Extraction Kits be used to process environmental or fecal samples for metagenomic studies?
- Can the Monarch HMW DNA Extraction Kits be used to isolate BAC DNA?
- How do you suggest homogenizing tougher tissues, like ear punches or muscle? Can a sonicator be used for tissue homogenization?
- Does the kit work with frozen tissue, tissue stored in preservatives (ethanol, DMSO-salt/DESS, or RNAlater®), FFPE samples, or OCT-preserved tissues?
- Do you see different fragmentation results when using the pestle versus the rotor-stator homogenizer for tissue homogenization?
- Do you have any PacBio Sequencing results from DNA extracted from tissue samples with the Monarch HMW DNA Extraction Kits?
- Does the Monarch HMW DNA Extraction workflow work for single cells?
- Can you use the DNA isolated with the Monarch HMW DNA Extraction Kits in Southern blots?
- Can the Monarch HMW DNA Extraction Kits be used to process nematodes?
- I am having trouble dissolving my HMW DNA after isolation. Do you have any tips on increasing the solubility?
- The enzymes in the kit were not stored at -20°C right away. Will they still work?
- There is a precipitate in the Proteinase K enzyme tube, is this normal?
- Which Monarch® columns and bead retainers can be used with which Monarch collection tube?
- I noticed the collection tube dimensions has changed slightly in the Monarch® HMW DNA Extraction Kit for Cells & Blood (NEB #T3050) and the Monarch HMW DNA Extraction Kit for Tissue (NEB #T3060). Will this impact performance?
- I notice that I'm applying force to fit the column into a collection tube. Should I be doing this? What collection tubes are compatible with columns?
Troubleshooting
Citations & Technical Literature
Citations
Additional Citations
Quality, Safety & Legal
Quality Assurance Statement
Quality Control tests are performed on each new lot of NEB product to meet the specifications designated for it. Specifications and individual lot data from the tests that are performed for this particular product can be found and downloaded on the Product Specification Sheet, Certificate of Analysis, data card or product manual. Further information regarding NEB product quality can be found here.Specification Change Notifications
Effective March 18,2024, collection tube dimensions modified and component numbers changed.
Effective March 14, 2024: Proteinase K, Molecular Biology Grade component number has changed with the concentration changing from 800U/ml to 20 mg/ml.
Specifications
The Specification sheet is a document that includes the storage temperature, shelf life and the specifications designated for the product. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]Certificate Of Analysis
The Certificate of Analysis (COA) is a signed document that includes the storage temperature, expiration date and quality controls for an individual lot. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]_[Lot Number]- T3060L_v1_10103689
- T3060S_v1_10103690
- T3060G_v1_10114355
- T3060L_v1_10118134
- T3060L_v2_10119971
- T3060S_v2_10119972
- T3060L_v2_10123959
- T3060G_v2_10119970
- T3060L_v2_10152960
- T3060S_v2_10152961
- T3060G_v2_10193018
- T3060L_v2_10193019
- T3060S_v2_10193020
- T3060L_v2_10216038
- T3060L_v3_10231530
- T3060G_v3_10231531
- T3060S_v3_10231545
Legal and Disclaimers
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.
New England Biolabs (NEB) is committed to practicing ethical science – we believe it is our job as researchers to ask the important questions that when answered will help preserve our quality of life and the world that we live in. However, this research should always be done in safe and ethical manner. Learn more.
Licenses
This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.
This product is covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc. For more information about commercial rights, please email us at gbd@neb.com While NEB develops and validates its products for various applications, the use of this product may require the buyer to obtain additional third party intellectual property rights for certain applications.
Other Products You May Be Interested In
-
Monarch® HMW DNA Extraction Kit for Cells & Blood
-

NEBNext® Companion Module for Oxford Nanopore Technologies® Ligation Sequencing
-

Monarch® Spin gDNA Extraction Kit
-
Proteinase K, Molecular Biology Grade
-
Monarch® RNase A
-

NEBNext® Ultra™ II DNA Library Prep Kit for Illumina®
-
Monarch® Pestle Set
-

Monarch® 2 ml Tubes
-
Monarch® DNA Capture Beads
-
Monarch® Bead Retainers
-
Monarch® gDNA Nuclei Prep & Lysis Buffer Pack
-

Monarch® RBC Lysis Buffer
-

Monarch® gDNA Elution Buffer II
-

Monarch® HMW gDNA Tissue Lysis Buffer
-
Monarch® Protein Separation Solution
-

Monarch® Precipitation Enhancer
The supporting documents available for this product can be downloaded below.